Sample Quality Control
Sample QC
We offer quality control of your nucleic acids as a stand alone service.
The RNA/DNA quality is crucial for microarray and next-generation sequencing experiments. The three quality criteria are concentration, purity and integrity.
1) Concentration
The concentration is measured using UV absorption at 260 nm with a spectrophotometer plate reader, the SpectroStar (BMG Labtech).
A Qubit is also an option for accurate concentration measurement.

2) Purity
The SpectroStar identifies possible sources of contamination.
Most protocols only mention the OD A260/A280 ratio but you will often see in the region of 230 nm a strong absorbing contaminant. Pure nucleic acids have a single peak with a maximum at 260 nm. A second peak at 230nm would indicate presence of contaminants within the sample. There are three sources of contaminations that produce peaks in the 220-230 nm region, these are proteins, chaotropic salts like guanidinium isothiocyanate and phenol.
In our opinion, it is important that both ratios, OD A260/A280 and OD A260/A230 should be in the 1.8- 2.1 region.
3) Integrity
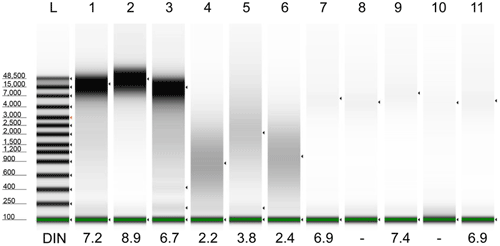
The RNA integrity is assessed on the Agilent Tapestation 4200 or the 2100 Bioanalyzer.
Total RNA run on either Agilent's instruments shows two distinct ribosomal peaks corresponding to either 18S and 28S for eukaryotic RNA and a relatively flat baseline between the 5S and 18S ribosomal peaks.
Agilent generates a RIN number (RNA Integrity Number (RIN) software algorithm) that takes the entire electropherogram into account to facilitate the assessment of the integrity of the total RNA preparation. This allows the classification of total RNA, based on a numbering system from 1 to 10, with 1 being the most degraded and 10 being the most intact. In this way, interpretation of an electropherogram and comparison between samples is facilitated.
For most RNA profiling by sequencing or array technologies, RINs higher than 8 is highly desirable. There are however solutions for working with samples of poorer quality samples (FFPE samples) with RINs in the region of 2-5. For example total RNA sequencing works well on degraded samples or using an array approach on the Affymetrix platform.

4) Sample shearing
We offer sample shearing on the Covaris platform. Please get in touch with us for more details on this service.
